SEREND 1.1
:: DESCRIPTION
SEREND is a semi-supervised learning method that uses a curated database of verified transcriptional factor-gene interactions, DNA sequence binding motifs, and a compendium of gene expression data in order to make thousands of new predictions about transcription factor-gene interactions, including whether the transcription factor activates or represses the gene.
::DEVELOPER
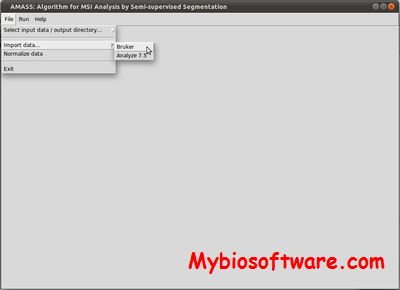
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux/MacOsX/ Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Ernst J, Beg QK, Kay KA, Balazsi G, Oltvai ZN, Bar-Joseph Z.
A Semi-Supervised Method for Predicting Transcription Factor-Gene Interactions in Escherichia coli.
PLoS Computational Biology 4: e1000044, 2008.