CNViewer 20111128
:: DESCRIPTION
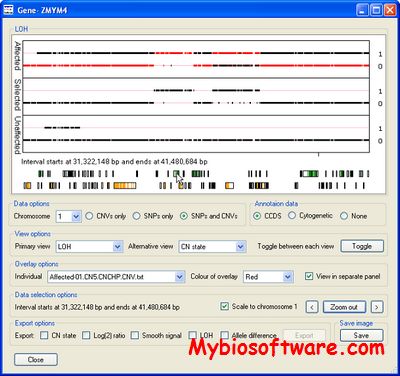
CNViewer aids the identification of regions of abnormal copy number variation that segregates with a disease phenotype in small pedigree.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
- Microsoft .NET framework version 2.0
:: DOWNLOAD
:: MORE INFORMATION
Citation
PLoS One. 2012;7(8):e43466. doi: 10.1371/journal.pone.0043466. Epub 2012 Aug 17.
Rapid visualisation of microarray copy number data for the detection of structural variations linked to a disease phenotype.
Carr IM, Diggle CP, Khan K, Inglehearn C, McKibbin M, Bonthron DT, Markham AF, Anwar R, Dobbie A, Pena SD, Ali M.