SBSI 1.4.5
:: DESCRIPTION
SBSI (Systems Biology Software Infrastructure) is a suite of software applications and libraries with the following aims:
- To provide access to parallelized algorithms for computationally challenging tasks such as parameter optimisation.
- To enable registered users to run these algorithms on their models and data, using HPC machines.
- To provide access for users with varying levels of computational modelling experience.
There are three main software components in SBSI: SBSINumerics, SBSIVisual and SBSIDispatcher.
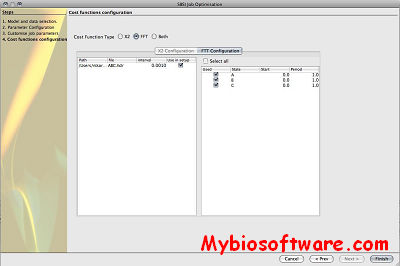
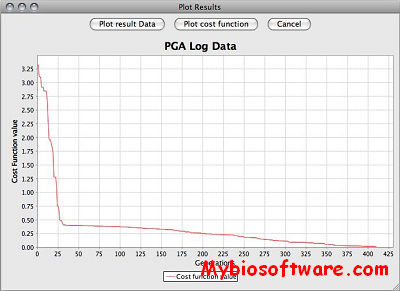
SBSINumerics contains a configurable, parallelized command line application for running parameter estimations.
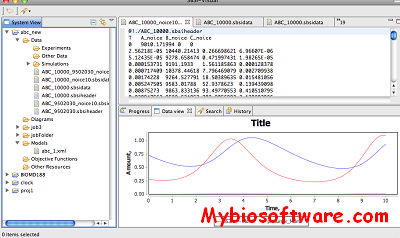
SBSIVisual is a desktop application, that provides easier access to SBSINumerics functionality for non-programmers, and can display results, run simulations, and interact with systems biology databases.
SBSIDispatcher is a middleware component, that can deploy and manage SBSINumerics procedures on backend servers.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / Mac OsX
:: DOWNLOAD
:: MORE INFORMATION
Citation
SBSI: an extensible distributed software infrastructure for parameter estimation in systems biology.
Adams R, Clark A, Yamaguchi A, Hanlon N, Tsorman N, Ali S, Lebedeva G, Goltsov A, Sorokin A, Akman OE, Troein C, Millar AJ, Goryanin I, Gilmore S.
Bioinformatics. 2013 Mar 1;29(5):664-665. Epub 2013 Jan 17.