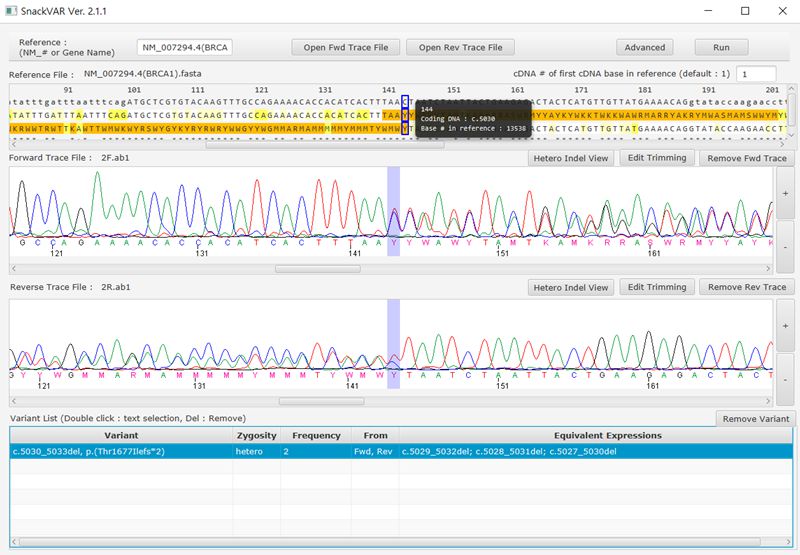
SnackVar v2.4.3
:: DESCRIPTION
SnackVar is a free software for Sanger sequencing analysis in clinical environment.It supports automatic detection of variants including SNVs and indel variants (homozygous and heterozygous).
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / MacOS
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Kim YG, Kim MJ, Lee JS, Lee JA, Song JY, Cho SI, Park SS, Seong MW.
SnackVar: An Open-Source Software for Sanger Sequencing Analysis Optimized for Clinical Use.
J Mol Diagn. 2021 Feb;23(2):140-148. doi: 10.1016/j.jmoldx.2020.11.001. Epub 2020 Nov 24. PMID: 33246077.

 NO
NO