RnaViz 2.0.3
:: DESCRIPTION
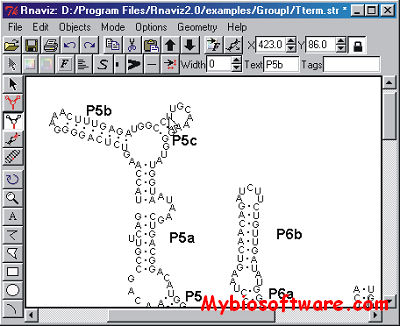
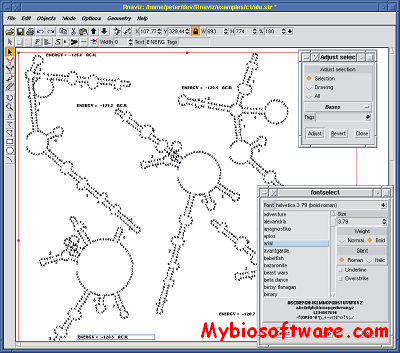
RnaViz is a user-friendly, portable, GUI program for producing publication-quality secondary structure drawings of RNA molecules. Drawings can be created starting from DCSE alignment files if they incorporate structure information or from mfold ct files. The layout of a structure can be changed easily. Display of special structural elements such as pseudo-knots or unformatted areas is possible. Sequences can be automatically numbered, and several other types of labels can be used to annotate particular bases or areas. Although the program does not try to produce an initially non-overlapping drawing, the layout of a properly positioned structure drawing can be applied to newly created drawing using skeleton files. In this way a range of similar structures can be drawn with a minimum of effort. Skeletons for several types of RNA molecule are included with the program.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / MacOsX / Linux
- Tcl/Tk Software
- Extral
- ClassyTcl/Tk
:: DOWNLOAD
:: MORE INFORMATION
Citation
Peter De Rijk, Jan Wuyts and Rupert De Wachter (2003)
RnaViz2: an improved representation of RNA secondary structure.
Bioinformatics 19(2): 299-300