SBML2LATEX 0.9.9
:: DESCRIPTION
SBML2LATEX is a tool to convert files in the System Biology Markup Language SBML) format into LATEX files.
::DEVELOPER
the Interfaculty Institute for Biomedical Informatics (IBMI)
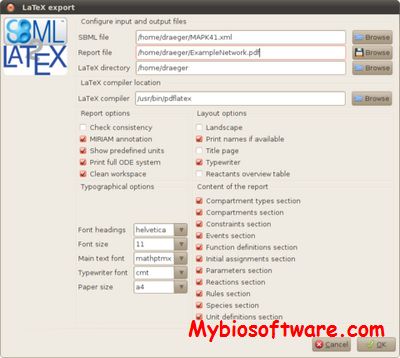
:: SCREENSHOTS
:: REQUIREMENTS
- Linux/ WIndows/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2009 Jun 1;25(11):1455-6. doi: 10.1093/bioinformatics/btp170. Epub 2009 Mar 23.
SBML2L(A)T(E)X: conversion of SBML files into human-readable reports.
Dräger A, Planatscher H, Motsou Wouamba D, Schröder A, Hucka M, Endler L, Golebiewski M, Müller W, Zell A.