ProtASR 2.2
:: DESCRIPTION
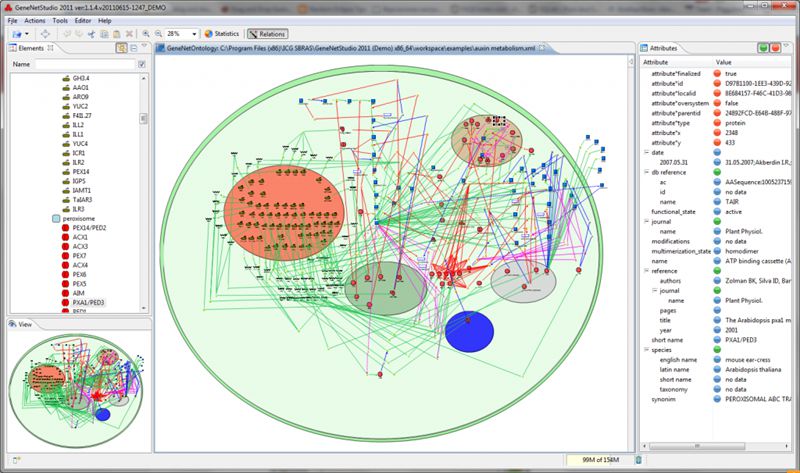
ProtASR is an evolutionary framework to reconstruct ancestral protein sequences accounting for structural constraints.
::DEVELOPER
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Arenas M, Weber CC, Liberles DA, Bastolla U.
ProtASR: An Evolutionary Framework for Ancestral Protein Reconstruction with Selection on Folding Stability.
Syst Biol. 2017 Nov 1;66(6):1054-1064. doi: 10.1093/sysbio/syw121. PMID: 28057858.