HapBound-GC and SHRUB-GC
:: DESCRIPTION
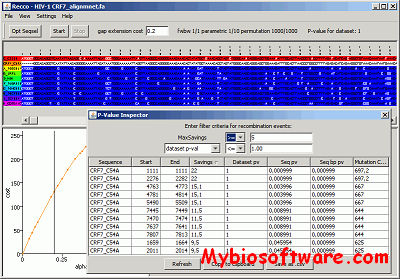
HapBound-GC and SHRUB-GC respectively compute lower and upper bounds on the minimum combined number of crossover and gene-conversion recombinations. SHRUB-GC constructs a graphical representation of evolutionary history involving coalescent, mutation, crossover and gene-conversion events.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux/Windows with Cygwin /MacOsX
:: DOWNLOAD
:: MORE INFORMATION
Citation
Song, Y.S., Ding, Z., Gusfield, D., Langley, C.H., and Wu, Y.
Algorithms to Distinguish the Role of Gene-Conversion from Single-Crossover Recombination in the Derivation of SNP Sequences
Lecture Notes in Computer Science 3909, (2006) 231-245.”