mlRho 2.9
:: DESCRIPTION
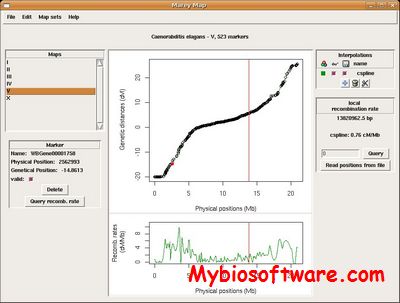
mlRho takes as input a file with assembled reads from a single diploid individual and returns maximum likelihood estimates of the population mutation rate, ![]() , the sequencing error
, the sequencing error ![]() , the zygosity correlation, and the population recombination rate,
, the zygosity correlation, and the population recombination rate, ![]()
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux/ MacOsX
- C Compiler
:: DOWNLOAD
:: MORE INFORMATION
Citation
B. Haubold, P. Pfaffelhuber, and M. Lynch.
mlRho: A program for estimating the population mutation and recombination rates from shotgun-sequenced diploid genomes.
Molecular Ecology, 19:277-284, 2010.