QUALITY 1.1.6
:: DESCRIPTION
The program QUALITY is a variant of the minimum Chi-squared (MC) method for limiting dilution assays, and for which he has demonstrated by simulation desirable properties of minimum variance (i.e., high precision) and minimum bias. Our method modifies the MC method to allow the user to specify the probabilities of a false negative and false positive PCR.
::DEVELOPER
Mullins Molecular Retrovirology Lab, University of Washington.
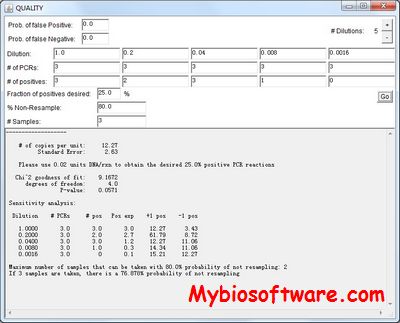
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
AIDS Res Hum Retroviruses. 1997 Jun 10;13(9):737-42.
Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays.
Rodrigo AG1, Goracke PC, Rowhanian K, Mullins JI.