PFP 2.3
:: DESCRIPTION
PFP or PFplus / PatchFinderPlus is a web-based tool for extracting and displaying continuous electrostatic positive patches on protein surfaces. The input required for PFplus is either a four letter PDB code or a protein coordinate file in PDB format, provided by the user. PFplus computes the continuum electrostatics potential and extracts the largest positive patch for each protein chain in the PDB file.
::DEVELOPER
Mandel-Gutfreund Lab, at the Technion.
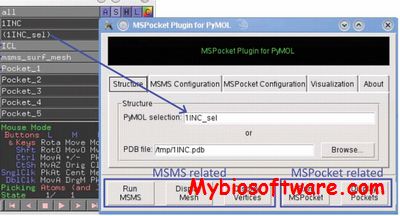
:: SCREENSHOTS
N/A
:: REQUIREMENTS
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
Shula Shazman, Gershon Celniker, Omer Haber, Fabian Glaser, Yael Mandel-Gutfreund (2007)
Patch Finder Plus (PFplus): A web server for extracting and displaying positive electrostatic patches on protein surfaces.
Nucleic Acids Res., 35:526-30.