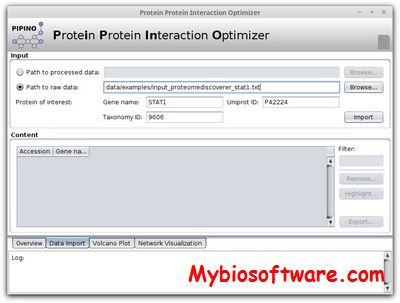
Strike
:: DESCRIPTION
Strike (String Kernel) is a program which classifies protein-protein interaction into “interacting” and “non-interacting” sets based solely on amino acid sequence information. The classification is made by applying the string kernel technique. Two proteins are classified “interacting” if they contain similar subsequences of amino acids.
::DEVELOPER
Nazar Zaki , Bioinformatics Laboratory, UAE University.
:: SCREENSHOTS
N/A
::REQUIREMENTS
:: DOWNLOAD
 Strike
Strike
:: MORE INFORMATION
Citation
Adv Exp Med Biol. 2011;696:263-70. doi: 10.1007/978-1-4419-7046-6_26.
Strike: a protein-protein interaction classification approach.
Zaki N, El-Hajj W, Kamel HM, Sibai F.