SCIP 1.0
:: DESCRIPTION
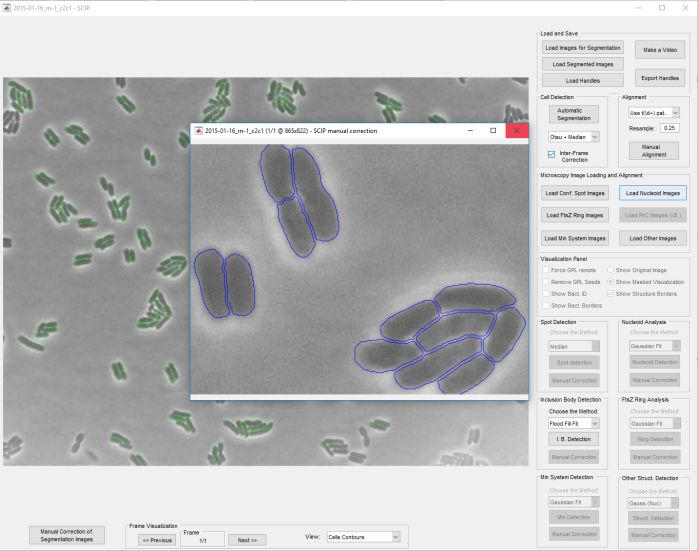
SCIP is an open source multi-channel, multi-process, time-lapse morphological and functional microscopy images analyser.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux/ MacOsX
- MatLab
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Martins L, Neeli-Venkata R, Oliveira SMD, Häkkinen A, Ribeiro AS, Fonseca JM.
SCIP: a single-cell image processor toolbox.
Bioinformatics. 2018 Dec 15;34(24):4318-4320. doi: 10.1093/bioinformatics/bty505. PMID: 29931314.