BatchPrimer3 1.0
:: DESCRIPTION
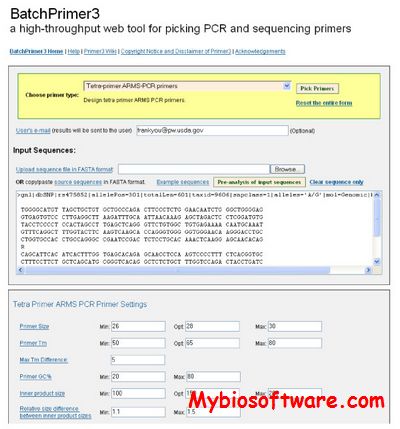
BatchPrimer3 is a comprehensive web primer design program using Primer3 core program as a major primer design engine to design different types of PCR primers and sequencing primers in a high-through manner. BatchPrimer3 allows users to design several types of primers including generic primers, hybridization oligos, SSR primers together with SSR detection, and SNP genotyping primers (including single-base extension primers, allele-specific primers, and tetra-primers for tetra-primer ARMS PCR), as well as DNA sequencing primers. A batch input of large number of sequences and a tab-delimited result output greatly facilitates rapid primer design and ordering process.
::DEVELOPER
Department of Plant Sciences, University of California
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Perl
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
BatchPrimer3: a high throughput web application for PCR and sequencing primer design.
You FM, Huo N, Gu YQ, Luo MC, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD.
BMC Bioinformatics. 2008 May 29;9:253.