MassShiftFinder 1.09
:: DESCRIPTION
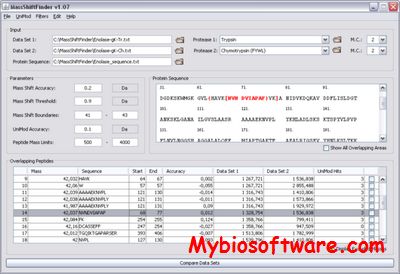
MassShiftFinder is a software tool for doing blind search using peptide mass fingerprints from two proteases with different cleavage specificities. The algorithm relies on overlapping peptides for the two proteases used, and can indicate both modifications and amino acid substitutions. The method can help restrict the area where the modification has occurred.
::DEVELOPER
The Proteomics Unit at the University of Bergen (PROBE)
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
BMC Res Notes. 2008 Dec 19;1:130.
Blind search for post-translational modifications and amino acid substitutions using peptide mass fingerprints from two proteases.
Barsnes H, Mikalsen SO, Eidhammer I.