Phylodendron 0.8d
:: DESCRIPTION
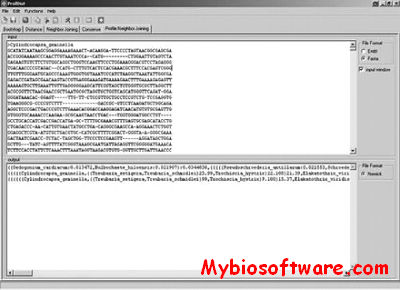
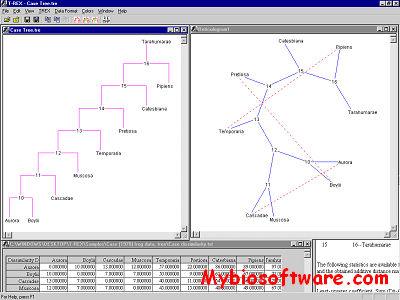
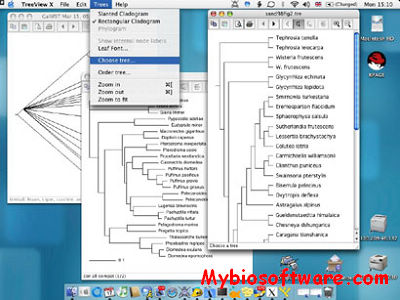
Phylodendron is a java software for drawing phylogenetic trees. Phylodendron is an application for drawing phylogenetic trees, used in evolutionary biology. It will read tree data in New Hampshire (Newick) format, then display graphical views of the phylogenetic tree. Various options allow you to modify, adorn and edit the tree. Standard application functions to save, print, edit and manage preferences are included. This program will not estimate nor produce the tree data. For that, software such as Phylip, Clustal W, and others may be used.
Phylodendron Online Version
::DEVELOPER
Don Gilbert at Indiana University
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux / Mac OsX
- Java
:: DOWNLOAD
 Phylodendron
Phylodendron
:: MORE INFORMATION
N/A