MOSAIC 1.1
:: DESCRIPTION
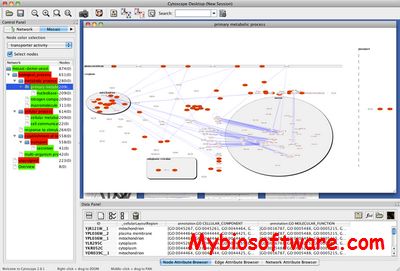
Mosaic performs network annotation and interactive partitioning driven by the Gene Ontology. The Mosaic algorithm works by first annotating the network with GO terms, followed by partitioning the network into a series of subnetworks based on the biological process annotation of nodes.
::DEVELOPER
the National Resource for Network Biology (NRNB)
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX /Windows
- Java
- Cytoscape
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Bioinformatics. 2012 Jul 15;28(14):1943-4. doi: 10.1093/bioinformatics/bts278. Epub 2012 May 9.
Mosaic: making biological sense of complex networks.
Zhang C1, Hanspers K, Kuchinsky A, Salomonis N, Xu D, Pico AR.