NPS 1.3.2
:: DESCRIPTION
NPS (Nucleosome Positioning from Sequencing) is a signal processing-based algorithm for identifying positioned nucleosomes fromsequencing experiments at the nucleosome level.The software is a python software package that can identify nucleosome positions given histone-modification ChIP-seq or nucleosome sequencing at the nucleosome level. NPS obtains continuous wave-form that represents the enrichment of histone modifications (or nucleosomes) by extending each tag (25nt, Solexa) to 150nt in the 3’ direction and taking the middle 75, and detects the positions of nucleosomes based on Laplacian of Gaussian (LOG) edge detection. The p value of each detection was estimated using Poisson approximation and the user can decide a cut-off for the final selection of nucleosome positions. In case of histone modification, the sequence tags are regrouped by different types of histone modification after nucleosome positioning and then the p-value of a particular histone modification at a positioned nucleosome was calculated based on the tag count of that histone modification in the nucleosome region using Poisson distribution, similar to the method mentioned above. The user also can select a cut-off of p value in histone modification assignment.
::DEVELOPER
X. Shirley Liu Lab
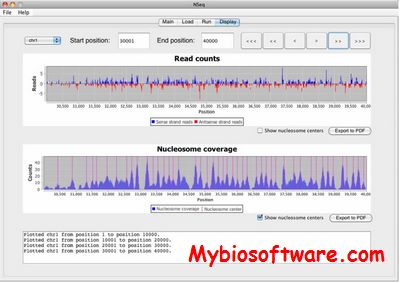
:: SCREENSHOTS
N/A
:: REQUIREMENTS
:: DOWNLOAD
 NPS
NPS
:: MORE INFORMATION
Citation:
Zhang Y, Shin H, Song JS, Lei Y, Liu XS.
Identifying Positioned Nucleosomes with Epigenetic Marks in Human from ChIP-Seq.
BMC Genomics 2008, 9:537.