PDBjViewer 4.5.5
:: DESCRIPTION
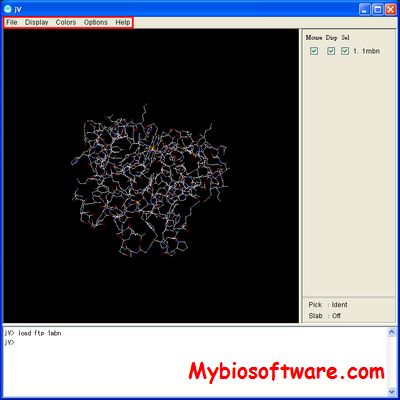
PDBjViewer (jV, for short) is a program to display molecular graphics of proteins and nucleic acids.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX / Windows
- Java
- JOGL
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2004 May 22;20(8):1329-30. Epub 2004 Feb 10.
eF-site and PDBjViewer: database and viewer for protein functional sites.
Kinoshita K, Nakamura H.