LACS
:: DESCRIPTION
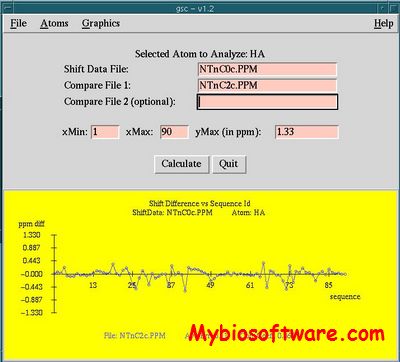
LACS ( linear analysis of chemical shifts) is a validation tool for protein backbone NMR chemical shift data. Based on the observation that secondary chemical shift (difference between the chemical shift of an amino acid and its random coil chemical shift) well indicate the local backbone geometry, LACS correlates the secondary chemical shifts of a nucleus to that of Ca-Cb of itself (referencing independent), thus avoiding any geometry or structure assumptions, to estimate referencing offsets and identify possible miss-assignments.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows
- Matlab
:: DOWNLOAD
:: MORE INFORMATION
Citation
Wang, L.; Eghbalnia, H. R.; Bahrami, A.; Markley, J. L.,
Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications,
J. Biomol. NMR 25, 32, 13-22.