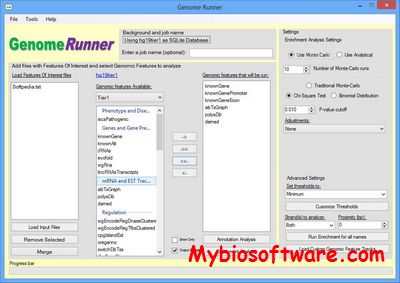
GenomeRunner 4.0
:: DESCRIPTION
GenomeRunner is a tool for automating genome exploration. It performs annotation and enrichment analyses of user-provided genomic regions (SNPs, ChIP-seq binding sites etc.) against >6,000 (human genome) epigenomic features available from the UCSC genome browser.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2012 Feb 1;28(3):419-20. doi: 10.1093/bioinformatics/btr666. Epub 2011 Dec 6.
GenomeRunner: automating genome exploration.
Dozmorov MG1, Cara LR, Giles CB, Wren JD.