mzMatch/PeakML 2.0
:: DESCRIPTION
mzMatch is a modular, open source and platform independent data processing pipeline for metabolomics LC/MS data written in the Java language. It was designed to provide small tools for the common processing tasks for LC/MS data. The mzMatch environment was based entirely on the PeakML file format and core library, which provides a common framework for all the tools.
::DEVELOPER
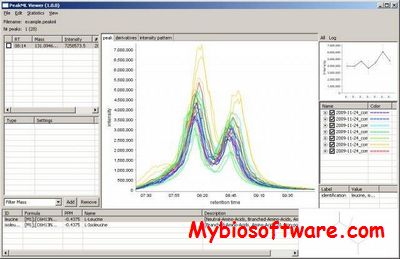
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java/ R
:: DOWNLOAD
:: MORE INFORMATION
Citation
PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis.
Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R.
Anal Chem. 2011 Apr 1;83(7):2786-93. doi: 10.1021/ac2000994