ProSeqViewer 1.0.42
:: DESCRIPTION
ProSeqViewer is a TypeScript library to visualize annotation on single sequences and multiple sequence alignments.
::DEVELOPER
:: SCREENSHOTS
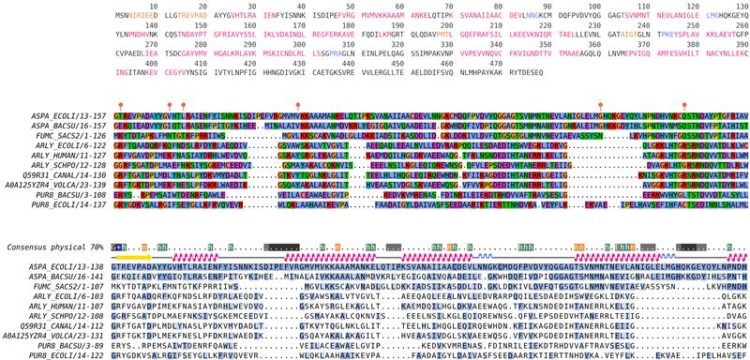
:: REQUIREMENTS
- Linux / Windows
- JS
:: DOWNLOAD
:: MORE INFORMATION
Citation:
Bevilacqua M, Paladin L, Tosatto SCE, Piovesan D.
ProSeqViewer: an interactive, responsive and efficient TypeScript library for visualization of sequences and alignments in web applications.
Bioinformatics. 2021 Nov 12:btab764. doi: 10.1093/bioinformatics/btab764. Epub ahead of print. PMID: 34788797.
