IMMP
:: DESCRIPTION
IMMP explores two approaches for the investigation of gene-miRNA relationships by integrating mRNA expression and protein expression data. It presents ranked miRNAs lists according to their effects on cancer development and constructed modules containing mRNAs, proteins, and miRNAs, in which these three molecular types are highly correlated.
::DEVELOPER
Data Mining & Computational Biology Lab
:: SCREENSHOTS
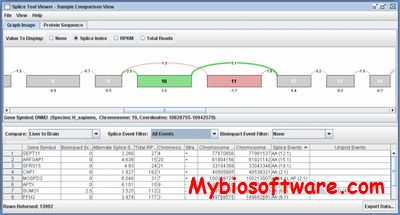
N/A
:: REQUIREMENTS
- Windows/Linux/MacOsX
- R
:: DOWNLOAD
:: MORE INFORMATION
Citation
Seo J, Jin D, Choi CH, Lee H.
Integration of MicroRNA, mRNA, and Protein Expression Data for the Identification of Cancer-Related MicroRNAs.
PLoS One. 2017 Jan 5;12(1):e0168412. doi: 10.1371/journal.pone.0168412. PMID: 28056026; PMCID: PMC5215789.