UCSF Chimera 1.14
:: DESCRIPTION
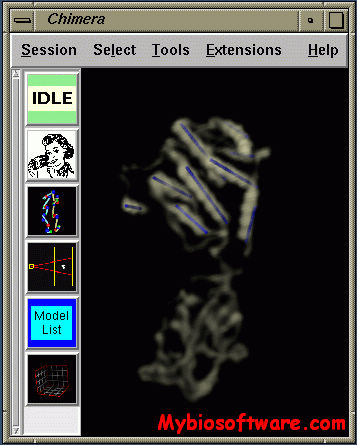
UCSF Chimera is a highly extensible program for interactive visualization and analysis of molecular structures and related data, including density maps, supramolecular assemblies, sequence alignments, docking results, trajectories, and conformational ensembles. High-quality images and animations can be generated. Chimera includes complete documentation and several tutorials, and can be downloaded free of charge for academic, government, non-profit, and personal use.
::DEVELOPER
the Resource for Biocomputing, Visualization, and Informatics (RBVI) at UCSF
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / MacOsX / Window
:: DOWNLOAD
:: MORE INFORMATION
Citation:
UCSF Chimera–a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.