NaviCell
:: DESCRIPTION
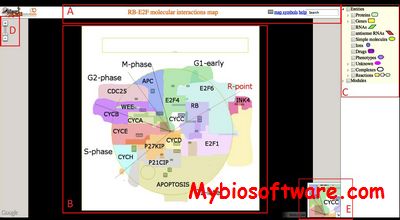
NaviCell is a web tool for exploring large maps of molecular interactions created in CellDesigner. The tool is characterized by a unique combination of three essential features: efficient map navigation based on Google maps engine, semantic zooming for viewing different levels of details on the map and an integrated blog for collecting the community curation feedbacks.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- any web browser
:: MORE INFORMATION
Citation
BMC Syst Biol. 2013 Oct 7;7:100. doi: 10.1186/1752-0509-7-100.
NaviCell: a web-based environment for navigation, curation and maintenance of large molecular interaction maps.
Kuperstein I1, Cohen DP, Pook S, Viara E, Calzone L, Barillot E, Zinovyev A.