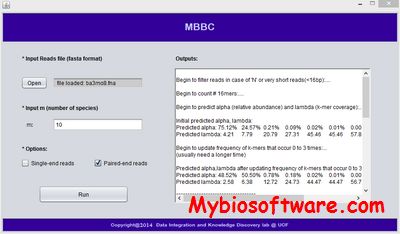
CLARK 1.2.6.1
:: DESCRIPTION
Clark is a novel approach to classify metagenomic reads at the species or genus level with high accuracy and high speed.
::DEVELOPER
Algorithms and Computational Biology Lab ,University of California
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Ounit R, Wanamaker S, Close TJ, Lonardi S,
CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers
BMC Genomics 2015, 16:236.