PhenoMeter 1.0
:: DESCRIPTION
The PhenoMeter is a metabolome database search tool that uses metabolite response patterns or metabolic phenotypes as queries, searching the MetabolomeExpress metabolomics database for significantly similar phenotypes and ranking them by similarity.
::DEVELOPER
:: SCREENSHOTS
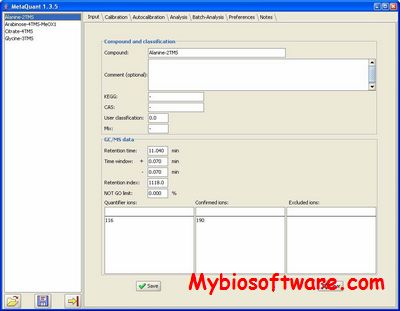
N/A
:: REQUIREMENTS
- Web browser
:: DOWNLOAD
 NO
NO
:: MORE INFORMATION
Citation
PhenoMeter: A Metabolome Database Search Tool Using Statistical Similarity Matching of Metabolic Phenotypes for High-Confidence Detection of Functional Links.
Carroll AJ, Zhang P, Whitehead L, Kaines S, Tcherkez G, Badger MR.
Front Bioeng Biotechnol. 2015 Jul 29;3:106. doi: 10.3389/fbioe.2015.00106.