metabosearch
:: DESCRIPTION
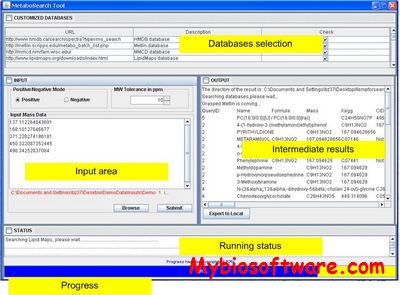
MetaboSearch performs mass-based metabolite search simultaneously against the four major metabolite databases: Human Metabolome DataBase (HMDB), Madison Metabolomics Consortium Database (MMCD), Metlin, and LipidMaps.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Linux / Windows/ MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
PLoS One. 2012;7(6):e40096. doi: 10.1371/journal.pone.0040096. Epub 2012 Jun 29.
MetaboSearch: tool for mass-based metabolite identification using multiple databases.
Zhou B1, Wang J, Ressom HW.