disentangler 20111001
:: DESCRIPTION
The disentangler is a visualization technique for linkage disequilibrium mapping and haplotype analysis of multiple multi-allelic genetic markers.
::DEVELOPER
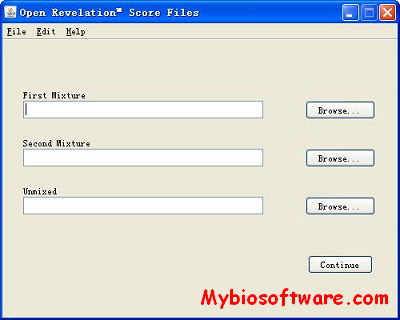
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux / MacOsX / Windows
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
N. Kumasaka, Y. Okada, A. Takahashi, M. Kubo, Y. Nakamura and N. Kamatani.
Disentangler: a visualization technique for linkage disequilibrium mapping using multi-allelic loci ;
(Abstract / Program #708F). Presented at the 12th International Congress of Human Genetics/61st Annual Meeting of The American Society of Human Genetics, October 14, 2011, Montreal, Canada.