LTR_STRUC
:: DESCRIPTION
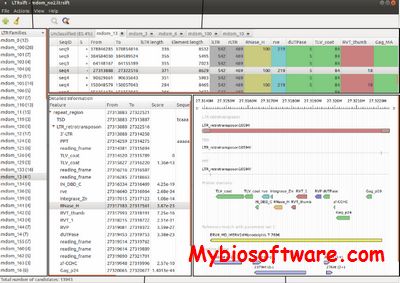
LTR_STRUC is a new data-mining program that scans nucleotide sequence files for LTR retrotransposons and analyzes any resulting hits. Input files can be in fasta or NCBI flat file format.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
 Contact with Vinay Mittal ( vinaykmittal@gatech.edu )
Contact with Vinay Mittal ( vinaykmittal@gatech.edu )
:: MORE INFORMATION
Citation:
McCarthy, E.M., and J.F. McDonald. 2003.
LTR_STRUC: A novel search and identification program for LTR retrotransposons.
Bioinformatics 19: 362-367