LINKAGE
:: DESCRIPTION
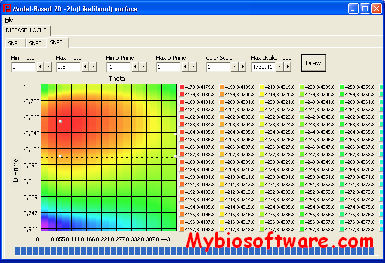
LINKAGE package is a series of programs for maximum likelihood estimation of recombination rates, calculation of lod score tables, and analysis of genetic risks. The analysis programs are divided into two groups. The first group can be used for general pedigrees with marker and disease loci. Programs in the second group are for three-generation families and codominant marker loci, and are primarily intended for the construction of genetic maps from data on reference families.
::DEVELOPER
Lab of Statistical Genetics, Rockefeller University
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows / Linux / MacOS
:: DOWNLOAD
:: MORE INFORMATION