LEVER
:: DESCRIPTION
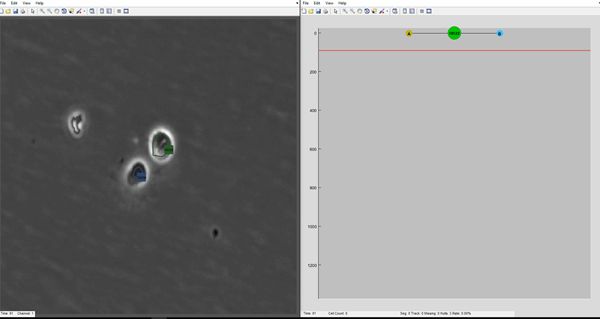
LEVER (Lineage Editing and Validation) is a collection of software tools for analyzing the development of proliferating (dividing) cells in time-lapse microscopy image sequences. LEVER includes algorithms for segmentation, tracking and lineaging. LEVER also includes a user interface that allows the segmentation, tracking and lineaging results to be validated, with any errors in the automated processing easily identified and quickly corrected
CloneView is an incredibly useful tool for visualizing the results from the LEVER program together with the image data.
::DEVELOPER
computational image sequence analysis
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
- MatLab
:: DOWNLOAD
:: MORE INFORMATION
Citation
LEVER: software tools for segmentation, tracking and lineaging of proliferating cells.
Winter M, Mankowski W, Wait E, Temple S, Cohen AR.
Bioinformatics. 2016 Jul 16. pii: btw406.