MultiGlycan 1.0
:: DESCRIPTION
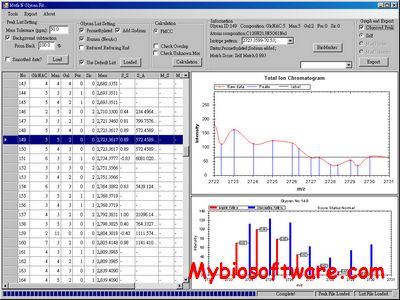
MultiGlycan helps user to gather glycan profile information from LC-MS Spectra. It also reports quantity of specific glycan composition.
::DEVELOPER
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2013 Jul 1;29(13):1706-7. doi: 10.1093/bioinformatics/btt190. Epub 2013 Apr 22.
Automated annotation and quantification of glycans using liquid chromatography-mass spectrometry.
Yu CY1, Mayampurath A, Hu Y, Zhou S, Mechref Y, Tang H.