pathClass 0.9.4
:: DESCRIPTION
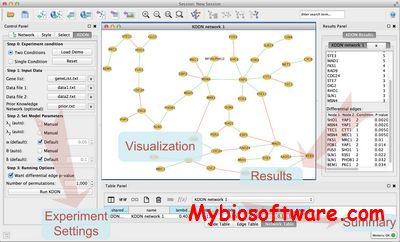
pathClass is a collection of classification methods that use information about feature connectivity in a biological network as an additional source of information. This additional knowledge is incorporated into the classification a priori. Several authors have shown that this approach significantly increases the classification performance.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows/Linux/MacOsX
- R
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2011 May 15;27(10):1442-3. doi: 10.1093/bioinformatics/btr157. Epub 2011 Mar 30.
pathClass: an R-package for integration of pathway knowledge into support vector machines for biomarker discovery.
Johannes M1, Fröhlich H, Sültmann H, Beissbarth T.