ICeE 2.10
:: DESCRIPTION
ICeE is a database with a Web-browser interface that allows you to enter experiments for the nematode C. elegans and to search efficiently in this database.
::DEVELOPER
the Ewbank lab
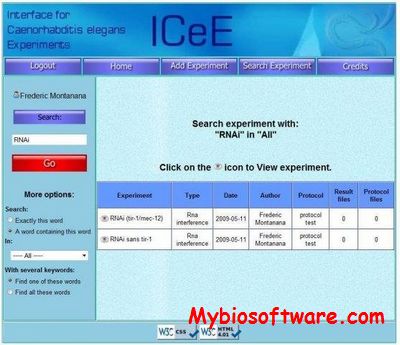
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/ Linux/ MacOsX
- Apache httpd
- MySQL
- PHP
:: DOWNLOAD
:: MORE INFORMATION