FRETraj v0.2.6
:: DESCRIPTION
FRETraj is a Python module for predicting FRET efficiencies by calculating multiple accessible-contact volumes (multi-ACV) to estimate donor and acceptor dye dynamics.
::DEVELOPER
FRETraj team
:: SCREENSHOTS
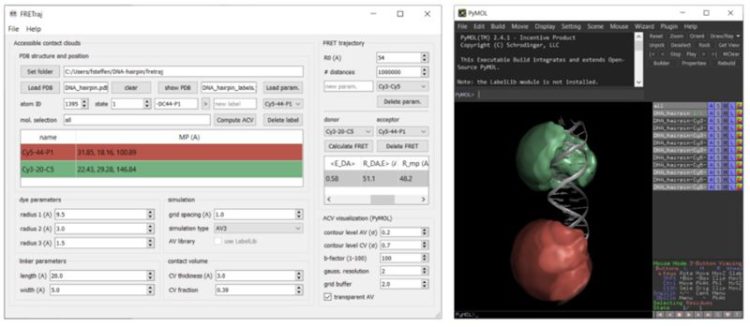
:: REQUIREMENTS
- Linux / Windows
- Python
- PyMOL
:: DOWNLOAD
:: MORE INFORMATION
Citation
Steffen FD, Sigel RKO, Börner R.
FRETraj: Integrating single-molecule spectroscopy with molecular dynamics.
Bioinformatics. 2021 Sep 3:btab615. doi: 10.1093/bioinformatics/btab615. Epub ahead of print. PMID: 34478493.


