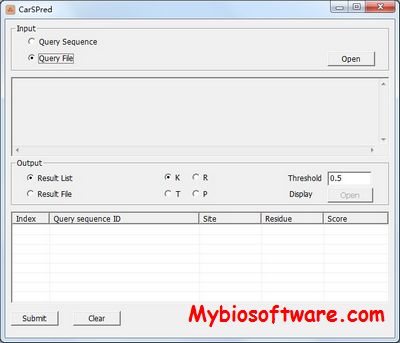
FpClass
:: DESCRIPTION
FpClass is a data mining-based method for proteome-wide PPI prediction.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
- Perl
:: DOWNLOAD
:: MORE INFORMATION
Citation
In silico prediction of physical protein interactions and characterization of interactome orphans.
Kotlyar M, Pastrello C, Pivetta F, Lo Sardo A, Cumbaa C, Li H, Naranian T, Niu Y, Ding Z, Vafaee F, Broackes-Carter F, Petschnigg J, Mills GB, Jurisicova A, Stagljar I, Maestro R, Jurisica I.
Nat Methods. 2015 Jan;12(1):79-84. doi: 10.1038/nmeth.3178.