PTest 20130115
:: DESCRIPTION
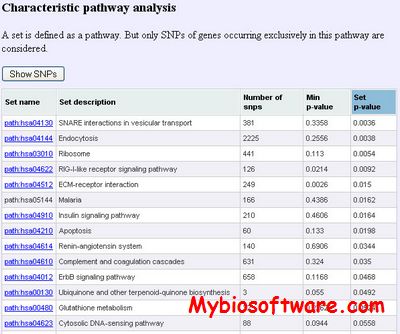
PTest (Partition test) is a software for case-control genetic association studies in human gene mapping. The PTest is based on single-SNP p-values resulting from an association test, for example, the chi-square test comparing genotype or allele frequencies in cases and controls.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Windows / Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Challenging false discovery rate: a partition test based on p values in human case-control association studies.
Ott J, Liu Z, Shen Y.
Hum Hered. 2012;74(1):45-50. doi: 10.1159/000343752. Epub 2012 Nov 13.