GeSTer
:: DESCRIPTION
GeSTer (GenomeScanner for Terminators) softwre first segregates the coding, upstream, and downstream regions based on the feature table entries of the genome sequence. Next it searches for palindromic sequences downstream of each gene (?20 to +270 nucleotides of the stop codon) without entering adjacent coding regions. The search is initiated at a G/C-rich (>50%) tetranucleotide, and a reverse complementary match is sought within the next 70 nucleotides. This defines the base of the stem, and the match is extended inward. Once a mismatch is encountered, all possible structures are computed allowing for different combinations of mismatches and gaps. The ΔGof formation of each of these structures was computed using the parameters from Turner et al. (22) and Jaeger et al. (23). Among all these structures, the one with the lowest ΔG was retained. Then the program moved to the next G/C-rich tetranucleotide and reinitiated the search.
::DEVELOPER
Ranjana Prakash,Indian Institute of Science, Bangalore. India
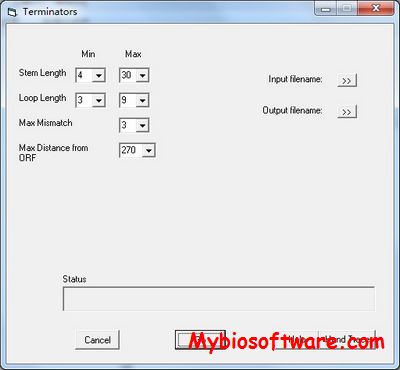
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION
Citation
Unniraman S, Prakash R, Nagaraja V:
Alternate paradigm for intrinsic transcription termination in eubacteria.
J Biol Chem 2001, 276:41850-55.