GIMM 3.8
:: DESCRIPTION
GIMM implements Bayesian infinite mixtures clustering procedures. The software consists of the R package gimmR and the self-standing software WinGimm.
:: DEVELOPER
Laboratory for Statistical Genomics, Univ. Cincinnati
:: SCREENSHOTS
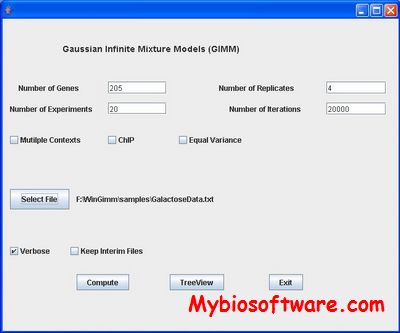
:: REQUIREMENTS
- Linux/MacOsX/Windows
- R Package
:: DOWNLOAD
 GIMM
GIMM
:: MORE INFORMATION
Citation:
Bioinformatics. 2006 Jul 15;22(14):1737-44. Epub 2006 May 18.
Context-specific infinite mixtures for clustering gene expression profiles across diverse microarray dataset.
Liu X, Sivaganesan S, Yeung KY, Guo J, Bumgarner RE, Medvedovic M.
BMC Bioinformatics. 2007 Aug 3;8:283.
Bayesian hierarchical model for transcriptional module discovery by jointly modeling gene expression and ChIP-chip data.
Liu X, Jessen WJ, Sivaganesan S, Aronow BJ, Medvedovic M.