GPViz 1.2.8
:: DESCRIPTION
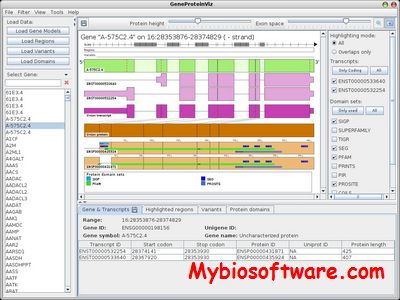
GPViz (GeneProteinViz) is a versatile Java-based software for dynamic gene-centered visualization of genomic regions and/or variants. User defined data can be loaded in common formats as resulting from analysis workflows used in sequencing applications and studied in the context of the gene, the corresponding transcript isoforms, proteins and their domains or other protein features. Both the genomic regions and variants can be also defined interactively.
::DEVELOPER
the Division of Bioinformatics, Innsbruck Medical University
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux /MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformatics. 2013 Sep 1;29(17):2195-6. doi: 10.1093/bioinformatics/btt354. Epub 2013 Jun 19.
GPViz: dynamic visualization of genomic regions and variants affecting protein domains.
Snajder R1, Trajanoski Z, Hackl H.