ITMO Genome Assembler 1.6.1
:: DESCRIPTION
ITMO Genome Assembler is a tool for de novo assembly of small and middle-sized genomes from reads, produced by Illumina/Solexa, Ion Torrent or Sanger sequencers.
::DEVELOPER
Computer Technologies Laboratory, National Research University of Information Technologies,
:: SCREENSHOTS
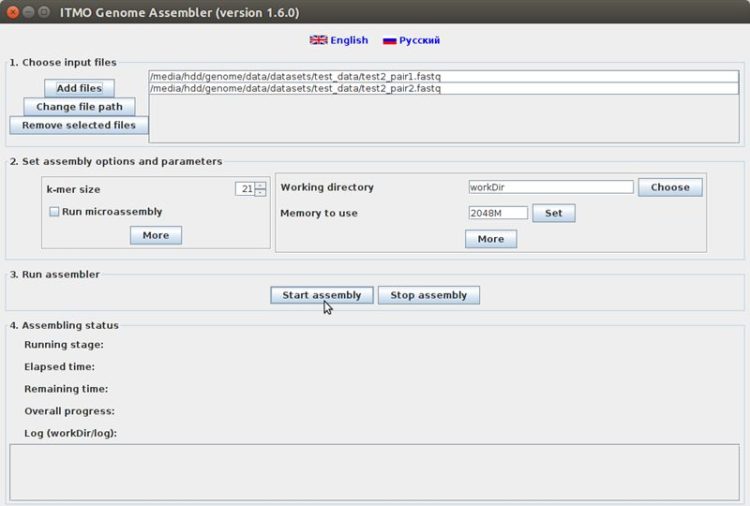
:: REQUIREMENTS
- Linux
- Java
:: MORE INFORMATION