Chromatin Cutter 1.00
:: DESCRIPTION
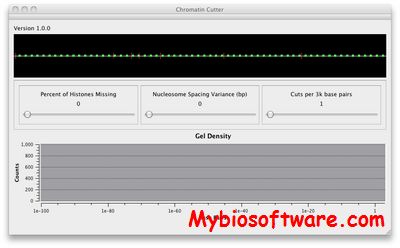
Chromatin Cutter simulates the distribution of DNA in a gel electrophoresis study of Yeast chromatin cut by enzymes. It lets you specify the fraction of nucleosomes that are unwrapped (wrapped ones have inaccessible DNA). It lets you specify the variance in the distribution of nucleosomes around their mean distance. It lets you specify the number of cuts per unit length (enzyme concentration). It plots the effects of these on the log number of base pairs, which matches the gel electrophoresis mass distribution.
::DEVELOPER
CISMM (Computer Integrated Systems for Microscopy and Manipulation)
:: SCREENSHOTS
:: REQUIREMENTS
- Windows
:: DOWNLOAD
:: MORE INFORMATION