G4ALL 1.1
:: DESCRIPTION
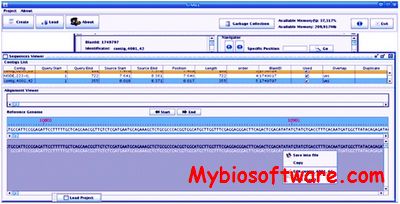
G4ALL is a stand-alone tool that facilitates the editing of the contigs generated by assembly process. G4ALL use mapping results produced by Blast or Mummer against a reference genome to generate a scaffold based on the overlap of the contigs after curation. Besides providing information on the gene products contained in each contig, obtained through a search of the available biological databases which can be inserted in G4ALL database.
::DEVELOPER
Dr. Rommel Ramos – rommelramos@ufpa.br
:: SCREENSHOTS
:: REQUIREMENTS
- Windows/Linux/MacOsX
- Java
:: DOWNLOAD
:: MORE INFORMATION
Citation
Bioinformation. 2013 Jun 29;9(11):599-604. doi: 10.6026/97320630009599. Print 2013.
Graphical contig analyzer for all sequencing platforms (G4ALL): a new stand-alone tool for finishing and draft generation of bacterial genomes.
Ramos RT1, Carneiro AR, Caracciolo PH, Azevedo V, Schneider MP, Barh D, Silva A.