FASPAD 20070415
:: DESCRIPTION
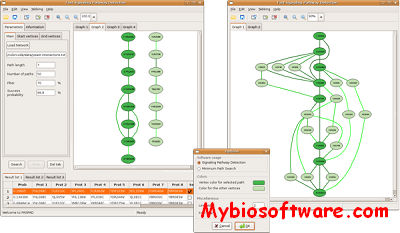
FASPAD (Fast Signaling Pathway Detection) is a tool for the detection of linear signaling pathways in protein interaction networks.Using recent algorithmic insights, it can solve the underlying NP-hard problem quite fast: for protein networks of typical size (several thousand nodes), pathway candidates of length up to 13 proteins can be found within seconds and with a 99.9% probability of optimality. FASPAD graphically displays all candidates that are found; for evaluation and comparison purposes, an overlay of several candidates and the surrounding network context can also be shown.
::DEVELOPER
Falk Hüffner and Thomas Zichner
:: SCREENSHOTS
:: REQUIREMENTS
- Windows / Linux
:: DOWNLOAD
 FASPAD for Linux ; for win ; Source Code
FASPAD for Linux ; for win ; Source Code
:: MORE INFORMATION
Citation
F. Hüffner, S. Wernicke, and T. Zichner.
Algorithm engineering for color-coding with applications to signaling pathway detection.
Algorithmica, 52(2):114–132, 2008