CoDiDi
:: DESCRIPTION
CoDiDi is program for estimating Gst, Hs and their correlation coefficient from marker data. It can be used to detect the presence or absence of mutational effect on the marker based genetic differentiation statistic Gst.
:: DEVELOPER
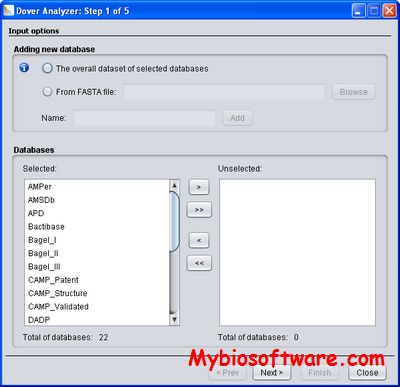
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux
:: DOWNLOAD
:: MORE INFORMATION
Citation
Wang J.
Does GST underestimate genetic differentiation from marker data?
Mol Ecol. 2015 Jul;24(14):3546-58. doi: 10.1111/mec.13204. Epub 2015 May 14. PMID: 25891752.



 NO
NO