AOP-helpFinder
:: DESCRIPTION
AOP-helpFinder is a web tool for identification and extraction of associations between stressors and biological events at various level of the biological organization (molecular initiating event (MIE), key event (KE), and adverse outcome (AO)).
::DEVELOPER
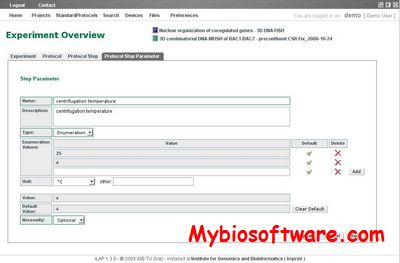
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
:: MORE INFORMATION
Citation
Jornod F, Jaylet T, Blaha L, Sarigiannis D, Tamisier L, Audouze K.
AOP-helpFinder webserver: a tool for comprehensive analysis of the literature to support adverse outcome pathways development.
Bioinformatics. 2021 Oct 30:btab750. doi: 10.1093/bioinformatics/btab750. Epub ahead of print. PMID: 34718414.