GenoFrag 2.1
:: DESCRIPTION
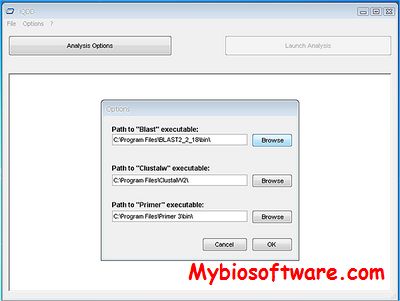
GenoFrag was developed for the analysis of Staphylococcus aureus genome plasticity by whole genome amplification in ~10 kb‐long fragments. A set of primers was generated from the genome sequence of S.aureus N315, employed here as a reference strain. Two subsets of primers were successfully used to amplify two portions of the N315 chromosome. This experimental validation demonstrates that GenoFrag is a robust and reliable tool for primer design and that whole genome PCR scanning can be envisaged for the analysis of genome diversity in S.aureus, one of the major public health concerns worldwide.
::DEVELOPER
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Linux/Windows/Unix/MacOsX
- Python
:: DOWNLOAD
:: MORE INFORMATION
Citation
Nouri Ben Zakour et. al.
GenoFrag: software to design primers optimized for whole genome scanning by long‐range PCR amplification
Nucl. Acids Res. (2004) 32 (1): 17-24