MiRGator 3.0
:: DESCRIPTION
miRGator aims to be the microRNA (miRNA) portal encompassing microRNA diversity, expression profiles, target relationships, and various supporting tools.
::DEVELOPER
Ewha Research Center for Systems Biology (ERCSB)
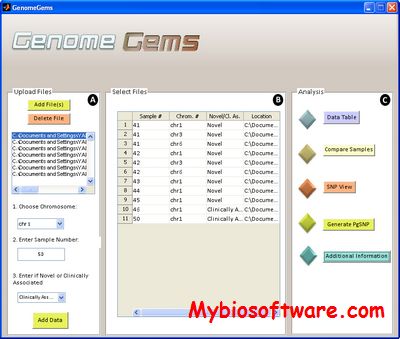
:: SCREENSHOTS
N/A
:: REQUIREMENTS
- Web Browser
:: DOWNLOAD
:: MORE INFORMATION
Citation
Nucleic Acids Res. 2013 Jan;41(Database issue):D252-7. doi: 10.1093/nar/gks1168. Epub 2012 Nov 27.
MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting.
Cho S1, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B, Kim VN, Kim W, Lee S.